Uncovering the NLR Family of Disease Resistance Genes in Cultivated Sweetpotato and Wild Relatives
Abstract
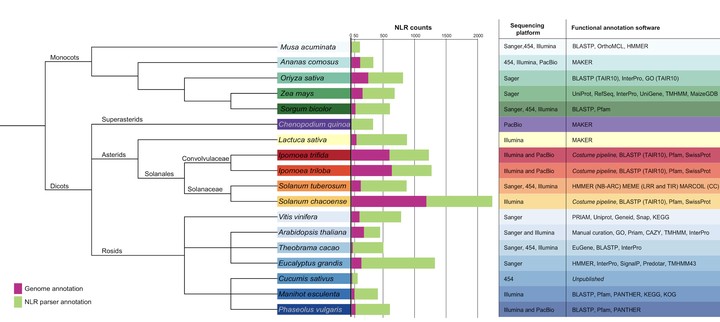
Sweetpotato (Ipomoea batatas) is an important staple crop cultivated on every continent except Antarctica. Awareness of sweetpotato nutritional benefits have led to an increase in consumption in the United States (US) and Europe. Despite continued implementation of disease management strategies, the fungal pathogen Ceratocystis fimbriata persists as a significant threat to the sweetpotato industry in the US. The presence of few breeding programs, limited knowledge of resistance, and the hexaploid nature of sweetpotato pose challenges to develop resistant lines. Plants possess an innate immune system with disease resistance ® genes encoding for proteins that recognize pathogen effectors during infection. An important class of resistance gene contains nucleotide-binding and leucine-rich repeat domains, NLRs. Here, we review state of the art knowledge of sweetpotato NLRs as well as novel methods to predict NLRs in plant genomes. Despite the availability of chemical control options, fungicide use is becoming limited due to changes in regulation and the evolution of pathogen resistance to chemicals. Deployment of host resistance represents a desirable tool to decrease crop losses due to plant pathogens. NLR gene annotations provide significant insight into the resistome and a first step towards identifying genes effective for control of major plant pathogens of sweetpotato.